In-silico identification of novel inhibitors for human Aurora kinase B form the ZINC database using molecular docking-based virtual screening
DOI:
https://doi.org/10.3897/rrpharmacology.8.82977Abstract
Introduction: Aurora kinase enzymes play critical functions in mammals. Aurora kinases are mitotic regulators that are involved in a variety of processes during cell division. The overexpression of these enzymes is associated with tumour formation and is symptomatic of clinical circumstances in cancer patients who have been diagnosed.
Materials and methods: The current study reports an in-silico virtual screening (VS) and molecular docking analysis of 2500 compounds retrieved from the ZINC database and five current clinical trial compounds against Aurora Kinase B using AutoDock Vina to identify potential inhibitors.
Results and discussion: The top six compounds that resulted from the screening were ZINC00190959, ZINC07889110, ZINC0088285, ZINC01404326, ZINC00882846 and ZINC08813187, which showed lower free energy of binding (FEB) against the target protein binding pocket. The FEB were as follows: -11.92, -11.85, -11.46, -11.33, -11.21 and -11.1 kcal/mol, using AutoDock, and -11.7, -11.5, -11.2, -11.0, -10.8 and -10.6 kcal/mol for AutoDock Vina, respectively. These findings were superior to those obtained with the co-crystallized ligand VX-680, with a -7.5 kcal/mol and the current clinical trial drug. Finally, using a VS and molecular docking approach, novel Aurora kinase B inhibitors were effectively identified from the ZINC database fulfilling the Lipinski rule of five with low FEB and functional molecular interactions with the target protein.
Conclusion: The findings suggest that the six compounds could be used as a potential agent for cancer treatments.
Graphical abstract: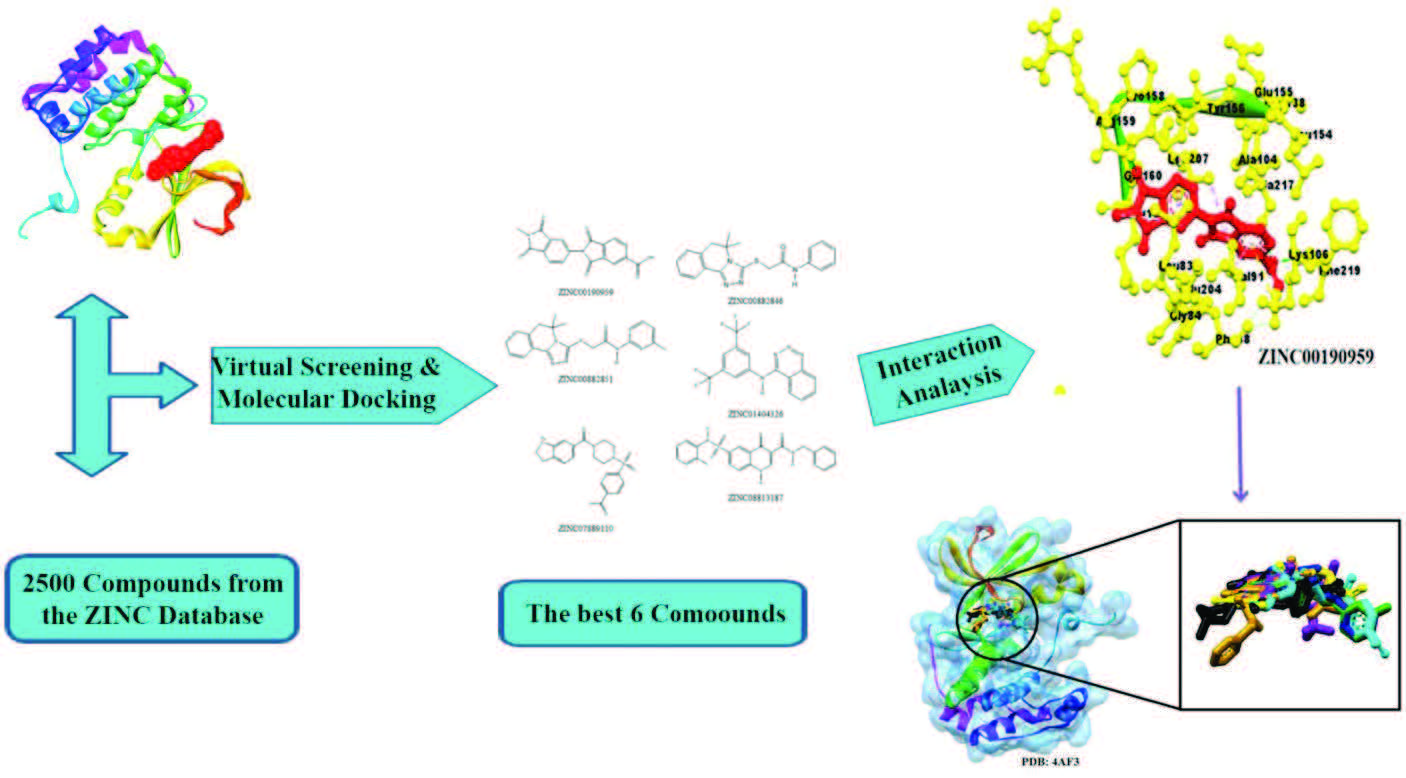
 Русский
Русский
 English
English

